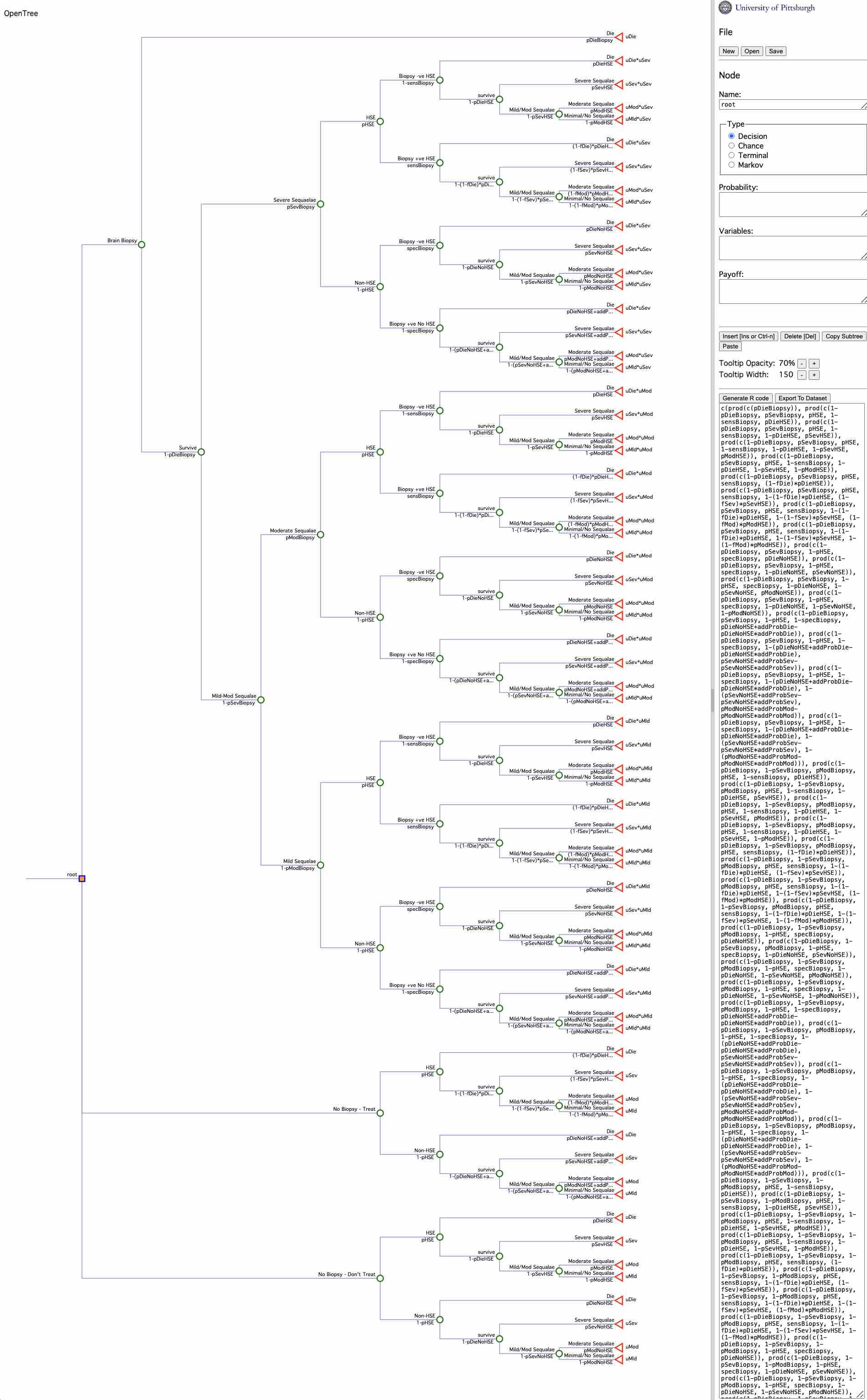
D3 Herpes Simplex Decision Tree Example Doubilet 1985
D3_dec_tree_Doubilet_1985_example.RmdThis is a relatively complex decision example involves treating herpes simplex encephalopathy and is published in Doubilet. The full model is described here:
Doubilet, P., Begg, C. B., Weinstein, M. C., Braun, P., & McNeil, B. J. (1985). Probabilistic sensitivity analysis using Monte Carlo simulation: a practical approach. Medical decision making, 5(2), 157-177.
It has the following features:
- How to control asymetrical decision branches - where the events are dependent on the decision
- How to make the probability of an event final_outcome based on the value of a prior (upstream) event
- How to make utility a function of a prior (upstream) event
rm(list = ls())
library(gmod)
#> Please note that active development of gmod is now moved to twig. Please install twig https://hjalal.github.io/twig/ and review the vignettes for the latest features.Setup model parameters as described in the paper
params <- list(
pDieBiopsy = 0.004,
pSevBiopsy = 0.01,
pModBiopsy = 0.03,
sensBiopsy = 0.95,
specBiopsy = 0.99,
pHSE = 0.4, #overall
pDieHSE = .7,
pSevHSE = .333,
pModHSE = .5,
fDie = .37,
fSev = .2,
fMod = .2,
pDieNoHSE = .18,
pSevNoHSE = .122,
pModNoHSE = .139,
addProbDie = .004,
addProbSev = .01,
addProbMod = .02,
uDie = 0,
uSev = 0.02,
uMod = .8,
uMld = 1
)This structure is meant to reproduce the tree structure as presented in Doubilet et al. However, their tree uses a binary biforcation, where each event can either be TRUE or FASLE. Below we simplify this tree structure by adding more final_outcomes to each decision branch. The outcomes are identical.
Binary event final_outcomes similar to Doubilet 1985
mygmod <- gmod() +
decisions("BrainBiopsy", "NoBiopsy_Treat", "NoBiopsy_NoTreat") +
#final_outcomes("DEAD","SEVSEQHSE","MODSEQHSE","MLDSEQHSE") +
event(name = "Biopsy",
scenarios = c(TRUE, FALSE),
probs = c(pBiopsy(decision), Inf),
outcomes = c("dieBiop", "HSE")) +
event(name = "dieBiop",
scenarios = c(TRUE, FALSE),
probs = c(pDieBiopsy, Inf),
outcomes = c("DEAD", "sevBiopSeq")) +
event(name = "sevBiopSeq",
scenarios = c(TRUE, FALSE),
probs = c(pSevBiopsy, Inf),
outcomes = c("HSE", "modBiopSeq")) +
event(name = "modBiopSeq",
scenarios = c(TRUE, FALSE),
probs = c(pModBiopsy, Inf),
outcomes = c("HSE", "HSE")) +
event(name = "HSE",
scenarios = c(TRUE, FALSE),
probs = c(pHSE, Inf),
outcomes = c("BiopAvail", "BiopAvail")) +
event(name = "BiopAvail",
scenarios = c(TRUE, FALSE),
probs = c(pBiopsy(decision), Inf),
outcomes = c("BiopRes", "die")) +
event(name = "BiopRes",
#scenarios = c("Positive", "Negative"),
scenarios = c(TRUE, FALSE),
probs = c(pBiopRes(HSE), Inf),
outcomes = c("die", "die")) +
event(name = "die",
scenarios = c(T, F),
probs = c(pEvent(HSE, decision, BiopRes, pDieHSE, pDieNoHSE, fDie, addProbDie), Inf),
outcomes = c("DEAD", "sevSeqHSE")) +
event(name = "sevSeqHSE",
scenarios = c(T, F),
probs = c(pEvent(HSE, decision, BiopRes, pSevHSE, pSevNoHSE, fSev, addProbSev), Inf),
outcomes = c("SEVSEQHSE", "modSeqHSE")) +
event(name = "modSeqHSE",
scenarios = c(T, F),
probs = c(pEvent(HSE, decision, BiopRes, pModHSE, pModNoHSE, fMod, addProbMod), Inf),
outcomes = c("MODSEQHSE", "MLDSEQHSE")) +
payoffs(util = util(decision, final_outcome, sevBiopSeq, modBiopSeq))
pEvent <- function(HSE, decision, BiopRes, pEventHSE, pEventNoHSE, fEvent, addProbEvent){
pEventRx <- if (HSE){
(1-fEvent)*pEventHSE
} else {
pEventNoHSE+addProbEvent-pEventNoHSE*addProbEvent
}
pEventNoRx <- if (HSE) pEventHSE else pEventNoHSE
if (decision=="NoBiopsy_NoTreat") pEventNoRx else
if (decision=="NoBiopsy_Treat") pEventRx else
if (decision=="BrainBiopsy")
if (BiopRes) pEventRx else pEventNoRx
}
pBiopsy <- function(decision){
(decision=="BrainBiopsy")
}
pBiopRes <- function(HSE){
HSE*sensBiopsy + (!HSE)*(1-specBiopsy)
}
util <- function(decision, final_outcome, sevBiopSeq, modBiopSeq){
if (decision=="BrainBiopsy"){
if (!is.na(sevBiopSeq) & sevBiopSeq){
uMult <- uSev
} else {
if (!is.na(modBiopSeq) & modBiopSeq){
uMult <- uMod
} else {
uMult <- uMld
}
}
} else {
uMult <- 1
}
# uMult <- (decision=="BrainBiopsy")*(sevBiopSeq*uSev + modBiopSeq*uMod + (!modBiopSeq)*uMld) +
# (decision!="BrainBiopsy")
#print(uMult)
switch(final_outcome,
"DEAD" = uDie,
"SEVSEQHSE" = uSev*uMult,
"MODSEQHSE" = uMod*uMult,
"MLDSEQHSE" = uMld*uMult)
}
model_struc <- gmod_build(mygmod)
model_struc
#> $decisions
#> [1] "BrainBiopsy" "NoBiopsy_Treat" "NoBiopsy_NoTreat"
#>
#> $n_decisions
#> [1] 3
#>
#> $n_events
#> [1] 10
#>
#> $final_outcomes
#> [1] "DEAD" "SEVSEQHSE" "MODSEQHSE" "MLDSEQHSE"
#>
#> $n_final_outcomes
#> [1] 4
#>
#> $events
#> [1] "Biopsy" "dieBiop" "sevBiopSeq" "modBiopSeq" "HSE"
#> [6] "BiopAvail" "BiopRes" "die" "sevSeqHSE" "modSeqHSE"
#>
#> $payoffs
#> $payoffs$util
#> util(decision, final_outcome, sevBiopSeq, modBiopSeq)
#>
#>
#> $payoff_names
#> [1] "util"
#>
#> $n_payoffs
#> [1] 1
#>
#> $final_outcome_formulae
#> # A tibble: 291 × 15
#> # Groups: decision, final_outcome [12]
#> decision final_outcome path_id probs Biopsy dieBiop sevBiopSeq modBiopSeq
#> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
#> 1 BrainBiopsy DEAD 1 (pBio… TRUE TRUE FALSE FALSE
#> 2 BrainBiopsy DEAD 2 (1-(p… FALSE FALSE FALSE FALSE
#> 3 BrainBiopsy DEAD 3 (pBio… TRUE FALSE TRUE FALSE
#> 4 BrainBiopsy DEAD 4 (pBio… TRUE FALSE FALSE TRUE
#> 5 BrainBiopsy DEAD 5 (pBio… TRUE FALSE FALSE FALSE
#> 6 BrainBiopsy DEAD 6 (1-(p… FALSE FALSE FALSE FALSE
#> 7 BrainBiopsy DEAD 7 (pBio… TRUE FALSE TRUE FALSE
#> 8 BrainBiopsy DEAD 8 (pBio… TRUE FALSE FALSE TRUE
#> 9 BrainBiopsy DEAD 9 (pBio… TRUE FALSE FALSE FALSE
#> 10 BrainBiopsy DEAD 10 (1-(p… FALSE FALSE FALSE FALSE
#> # ℹ 281 more rows
#> # ℹ 7 more variables: HSE <chr>, BiopAvail <chr>, BiopRes <chr>, die <chr>,
#> # sevSeqHSE <chr>, modSeqHSE <chr>, util <chr>
#>
#> $summary_formulae
#> # A tibble: 3 × 2
#> decision util
#> <chr> <chr>
#> 1 BrainBiopsy (pBiopsy('BrainBiopsy'))*(pDieBiopsy)*util('BrainBiopsy', 'D…
#> 2 NoBiopsy_NoTreat (pBiopsy('NoBiopsy_NoTreat'))*(pDieBiopsy)*util('NoBiopsy_No…
#> 3 NoBiopsy_Treat (pBiopsy('NoBiopsy_Treat'))*(pDieBiopsy)*util('NoBiopsy_Trea…
#>
#> attr(,"class")
#> [1] "gmod_decision"
gmod_gen_model_function(model_struc, model_function_name = "doubilet_1985_model")
#> Warning in gmod_gen_model_function.gmod_decision(model_struc, model_function_name = "doubilet_1985_model"): Model function doubilet_1985_model is generated. It can be run by calling it directly:
#> doubilet_1985_model(params)
#> $decisions
#> [1] "BrainBiopsy" "NoBiopsy_Treat" "NoBiopsy_NoTreat"
#>
#> $n_decisions
#> [1] 3
#>
#> $n_events
#> [1] 10
#>
#> $final_outcomes
#> [1] "DEAD" "SEVSEQHSE" "MODSEQHSE" "MLDSEQHSE"
#>
#> $n_final_outcomes
#> [1] 4
#>
#> $events
#> [1] "Biopsy" "dieBiop" "sevBiopSeq" "modBiopSeq" "HSE"
#> [6] "BiopAvail" "BiopRes" "die" "sevSeqHSE" "modSeqHSE"
#>
#> $payoffs
#> $payoffs$util
#> util(decision, final_outcome, sevBiopSeq, modBiopSeq)
#>
#>
#> $payoff_names
#> [1] "util"
#>
#> $n_payoffs
#> [1] 1
#>
#> $final_outcome_formulae
#> # A tibble: 291 × 15
#> # Groups: decision, final_outcome [12]
#> decision final_outcome path_id probs Biopsy dieBiop sevBiopSeq modBiopSeq
#> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
#> 1 BrainBiopsy DEAD 1 (pBio… TRUE TRUE FALSE FALSE
#> 2 BrainBiopsy DEAD 2 (1-(p… FALSE FALSE FALSE FALSE
#> 3 BrainBiopsy DEAD 3 (pBio… TRUE FALSE TRUE FALSE
#> 4 BrainBiopsy DEAD 4 (pBio… TRUE FALSE FALSE TRUE
#> 5 BrainBiopsy DEAD 5 (pBio… TRUE FALSE FALSE FALSE
#> 6 BrainBiopsy DEAD 6 (1-(p… FALSE FALSE FALSE FALSE
#> 7 BrainBiopsy DEAD 7 (pBio… TRUE FALSE TRUE FALSE
#> 8 BrainBiopsy DEAD 8 (pBio… TRUE FALSE FALSE TRUE
#> 9 BrainBiopsy DEAD 9 (pBio… TRUE FALSE FALSE FALSE
#> 10 BrainBiopsy DEAD 10 (1-(p… FALSE FALSE FALSE FALSE
#> # ℹ 281 more rows
#> # ℹ 7 more variables: HSE <chr>, BiopAvail <chr>, BiopRes <chr>, die <chr>,
#> # sevSeqHSE <chr>, modSeqHSE <chr>, util <chr>
#>
#> $summary_formulae
#> # A tibble: 3 × 2
#> decision util
#> <chr> <chr>
#> 1 BrainBiopsy (pBiopsy('BrainBiopsy'))*(pDieBiopsy)*util('BrainBiopsy', 'D…
#> 2 NoBiopsy_NoTreat (pBiopsy('NoBiopsy_NoTreat'))*(pDieBiopsy)*util('NoBiopsy_No…
#> 3 NoBiopsy_Treat (pBiopsy('NoBiopsy_Treat'))*(pDieBiopsy)*util('NoBiopsy_Trea…
#>
#> attr(,"class")
#> [1] "gmod_decision"
doubilet_1985_model(params)
#> util
#> BrainBiopsy 0.5580326
#> NoBiopsy_NoTreat 0.4940027
#> NoBiopsy_Treat 0.5660208with simplification - where paths that generate 0 prob are removed
model_struc <- gmod_build(mygmod, params = params, simplify = TRUE)
model_struc
#> $decisions
#> [1] "BrainBiopsy" "NoBiopsy_Treat" "NoBiopsy_NoTreat"
#>
#> $n_decisions
#> [1] 3
#>
#> $n_events
#> [1] 10
#>
#> $final_outcomes
#> [1] "DEAD" "SEVSEQHSE" "MODSEQHSE" "MLDSEQHSE"
#>
#> $n_final_outcomes
#> [1] 4
#>
#> $events
#> [1] "Biopsy" "dieBiop" "sevBiopSeq" "modBiopSeq" "HSE"
#> [6] "BiopAvail" "BiopRes" "die" "sevSeqHSE" "modSeqHSE"
#>
#> $payoffs
#> $payoffs$util
#> util(decision, final_outcome, sevBiopSeq, modBiopSeq)
#>
#>
#> $payoff_names
#> [1] "util"
#>
#> $n_payoffs
#> [1] 1
#>
#> $final_outcome_formulae
#> # A tibble: 291 × 15
#> # Groups: decision, final_outcome [12]
#> decision final_outcome path_id probs Biopsy dieBiop sevBiopSeq modBiopSeq
#> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
#> 1 BrainBiopsy DEAD 1 (pBio… TRUE TRUE FALSE FALSE
#> 2 BrainBiopsy DEAD 2 (1-(p… FALSE FALSE FALSE FALSE
#> 3 BrainBiopsy DEAD 3 (pBio… TRUE FALSE TRUE FALSE
#> 4 BrainBiopsy DEAD 4 (pBio… TRUE FALSE FALSE TRUE
#> 5 BrainBiopsy DEAD 5 (pBio… TRUE FALSE FALSE FALSE
#> 6 BrainBiopsy DEAD 6 (1-(p… FALSE FALSE FALSE FALSE
#> 7 BrainBiopsy DEAD 7 (pBio… TRUE FALSE TRUE FALSE
#> 8 BrainBiopsy DEAD 8 (pBio… TRUE FALSE FALSE TRUE
#> 9 BrainBiopsy DEAD 9 (pBio… TRUE FALSE FALSE FALSE
#> 10 BrainBiopsy DEAD 10 (1-(p… FALSE FALSE FALSE FALSE
#> # ℹ 281 more rows
#> # ℹ 7 more variables: HSE <chr>, BiopAvail <chr>, BiopRes <chr>, die <chr>,
#> # sevSeqHSE <chr>, modSeqHSE <chr>, util <chr>
#>
#> $summary_formulae
#> # A tibble: 3 × 2
#> decision util
#> <chr> <chr>
#> 1 BrainBiopsy (pBiopsy('BrainBiopsy'))*(pDieBiopsy)*util('BrainBiopsy', 'D…
#> 2 NoBiopsy_NoTreat (1-(pBiopsy('NoBiopsy_NoTreat')))*(pHSE)*(1-(pBiopsy('NoBiop…
#> 3 NoBiopsy_Treat (1-(pBiopsy('NoBiopsy_Treat')))*(pHSE)*(1-(pBiopsy('NoBiopsy…
#>
#> attr(,"class")
#> [1] "gmod_decision"
gmod_gen_model_function(model_struc, model_function_name = "doubilet_1985_model")
#> Warning in gmod_gen_model_function.gmod_decision(model_struc, model_function_name = "doubilet_1985_model"): Model function doubilet_1985_model is generated. It can be run by calling it directly:
#> doubilet_1985_model(params)
#> $decisions
#> [1] "BrainBiopsy" "NoBiopsy_Treat" "NoBiopsy_NoTreat"
#>
#> $n_decisions
#> [1] 3
#>
#> $n_events
#> [1] 10
#>
#> $final_outcomes
#> [1] "DEAD" "SEVSEQHSE" "MODSEQHSE" "MLDSEQHSE"
#>
#> $n_final_outcomes
#> [1] 4
#>
#> $events
#> [1] "Biopsy" "dieBiop" "sevBiopSeq" "modBiopSeq" "HSE"
#> [6] "BiopAvail" "BiopRes" "die" "sevSeqHSE" "modSeqHSE"
#>
#> $payoffs
#> $payoffs$util
#> util(decision, final_outcome, sevBiopSeq, modBiopSeq)
#>
#>
#> $payoff_names
#> [1] "util"
#>
#> $n_payoffs
#> [1] 1
#>
#> $final_outcome_formulae
#> # A tibble: 291 × 15
#> # Groups: decision, final_outcome [12]
#> decision final_outcome path_id probs Biopsy dieBiop sevBiopSeq modBiopSeq
#> <chr> <chr> <dbl> <chr> <chr> <chr> <chr> <chr>
#> 1 BrainBiopsy DEAD 1 (pBio… TRUE TRUE FALSE FALSE
#> 2 BrainBiopsy DEAD 2 (1-(p… FALSE FALSE FALSE FALSE
#> 3 BrainBiopsy DEAD 3 (pBio… TRUE FALSE TRUE FALSE
#> 4 BrainBiopsy DEAD 4 (pBio… TRUE FALSE FALSE TRUE
#> 5 BrainBiopsy DEAD 5 (pBio… TRUE FALSE FALSE FALSE
#> 6 BrainBiopsy DEAD 6 (1-(p… FALSE FALSE FALSE FALSE
#> 7 BrainBiopsy DEAD 7 (pBio… TRUE FALSE TRUE FALSE
#> 8 BrainBiopsy DEAD 8 (pBio… TRUE FALSE FALSE TRUE
#> 9 BrainBiopsy DEAD 9 (pBio… TRUE FALSE FALSE FALSE
#> 10 BrainBiopsy DEAD 10 (1-(p… FALSE FALSE FALSE FALSE
#> # ℹ 281 more rows
#> # ℹ 7 more variables: HSE <chr>, BiopAvail <chr>, BiopRes <chr>, die <chr>,
#> # sevSeqHSE <chr>, modSeqHSE <chr>, util <chr>
#>
#> $summary_formulae
#> # A tibble: 3 × 2
#> decision util
#> <chr> <chr>
#> 1 BrainBiopsy (pBiopsy('BrainBiopsy'))*(pDieBiopsy)*util('BrainBiopsy', 'D…
#> 2 NoBiopsy_NoTreat (pBiopsy('NoBiopsy_NoTreat'))*(pDieBiopsy)*util('NoBiopsy_No…
#> 3 NoBiopsy_Treat (pBiopsy('NoBiopsy_Treat'))*(pDieBiopsy)*util('NoBiopsy_Trea…
#>
#> attr(,"class")
#> [1] "gmod_decision"
doubilet_1985_model(params)
#> util
#> BrainBiopsy 0.5580326
#> NoBiopsy_NoTreat 0.4940027
#> NoBiopsy_Treat 0.5660208